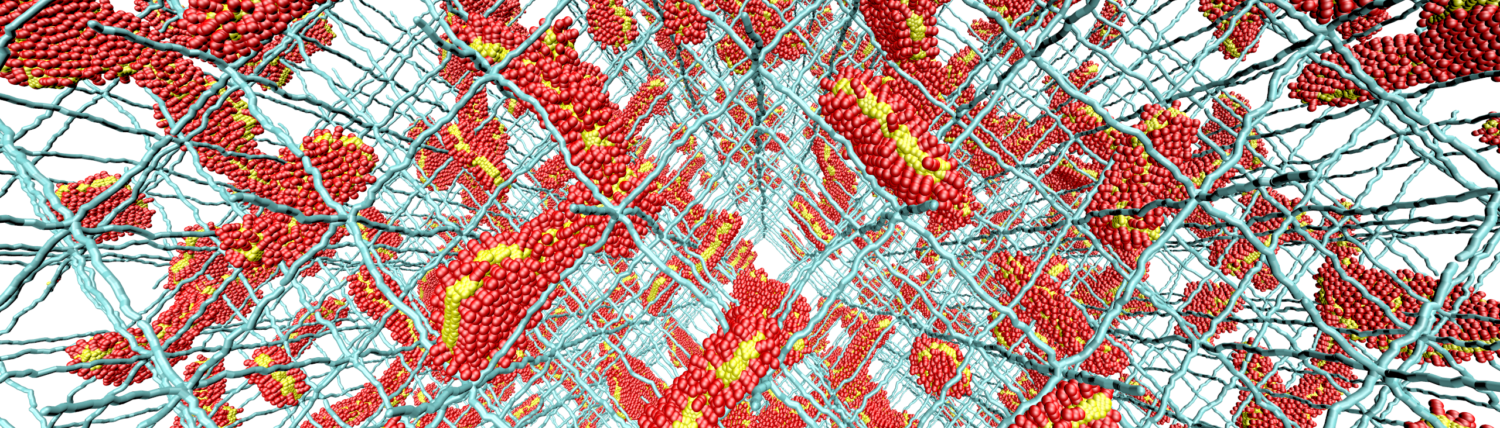
What is common between DNA carrying all the genetic information in your cells and plastic shopping bags ? Both of these “materials” are composed of long molecular chains that we refer to as polymers. Polymers, whether as constituents of biological or a human-made systems, can share common properties; they can respond to external stimuli, such as mechanical forces, electric fields, flows, or alteration in pH levels or temperature/concentration. They can also transiently adapt geometrical constraints. While all this versatility of polymeric soft-matter, on one hand, provides unprecedented survival and evolutionary capabilities for life (e.g., compaction of meters-long DNA in a protective nuclear shell), on the other hand, create new opportunities to design next-generation materials and pharmaceutical solutions.
Research in our lab focuses on fundamental problems related to polymers in biological and materials-science systems, such as DNA organization and hydrogels, on which we have currently either none or limited understanding. Specifically, we study the chromosome organization in cells and the potential role of protein-DNA interaction kinetics on this 3d organization . We are also interested in stimuli-responsive properties of polyelectrolyte hydrogels and polymer brushes. In our investigations, we use Molecular Dynamics (MD) simulations both on atomistic and coarse-grained levels, along with analytical tools of statistical mechanics.
Research Highlights
- Concentration-dependent off-rates of DNA-binding proteins in bacterial confinement
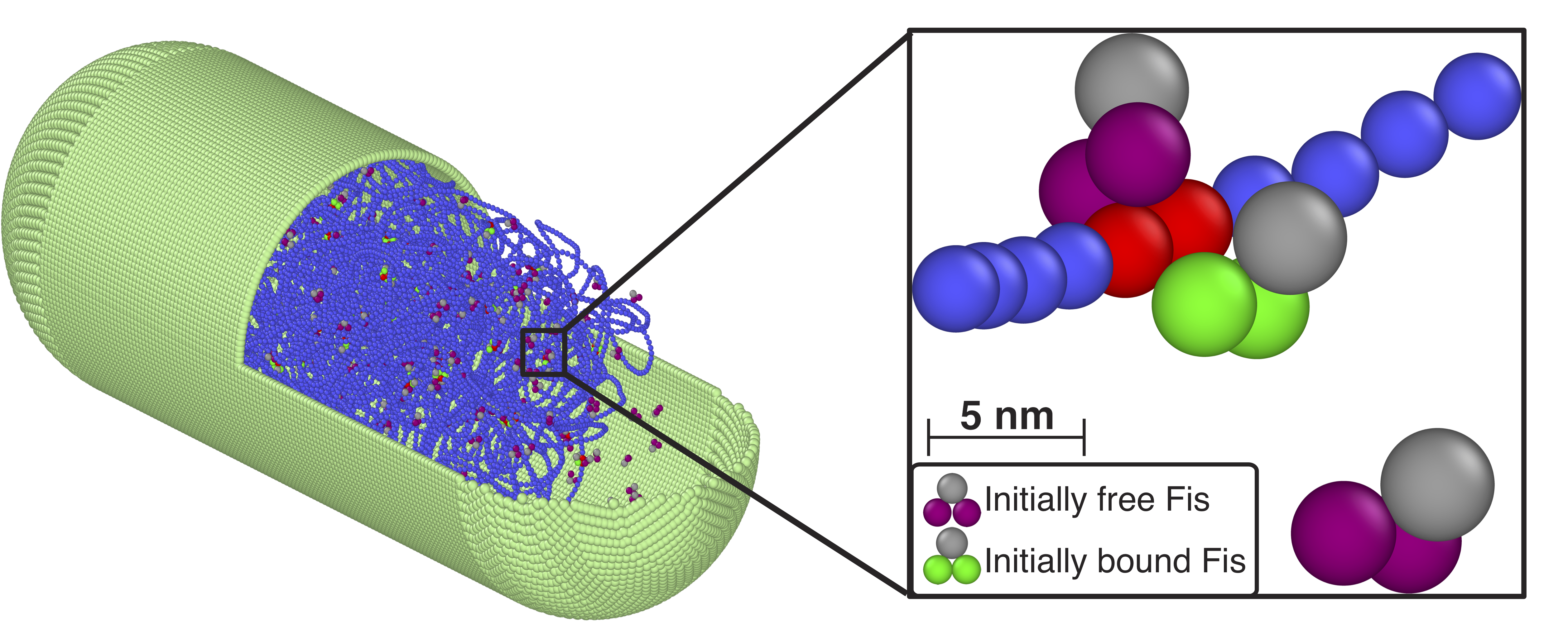
Almost all cellular functions from replication to transcription are controlled by the transient interactions between DNA and DNA-binding proteins. By using MD simulations of large-scale bacterial model, in which long DNA chain can interact with dimeric transcription factor proteins such as Fis or HU, we show that protein concentration influences the off-rates of the DNA-bound proteins. These proteins can also structurally change the chromosome compactions and further alter the off-rates by creating DNA-protein complexes and highly-collapsed regions of DNA. Read more on Biophysical J.
 |
 |
- Inhomogeneous chromatin flexibility contributes to the nuclear organization in eukaryotic cells
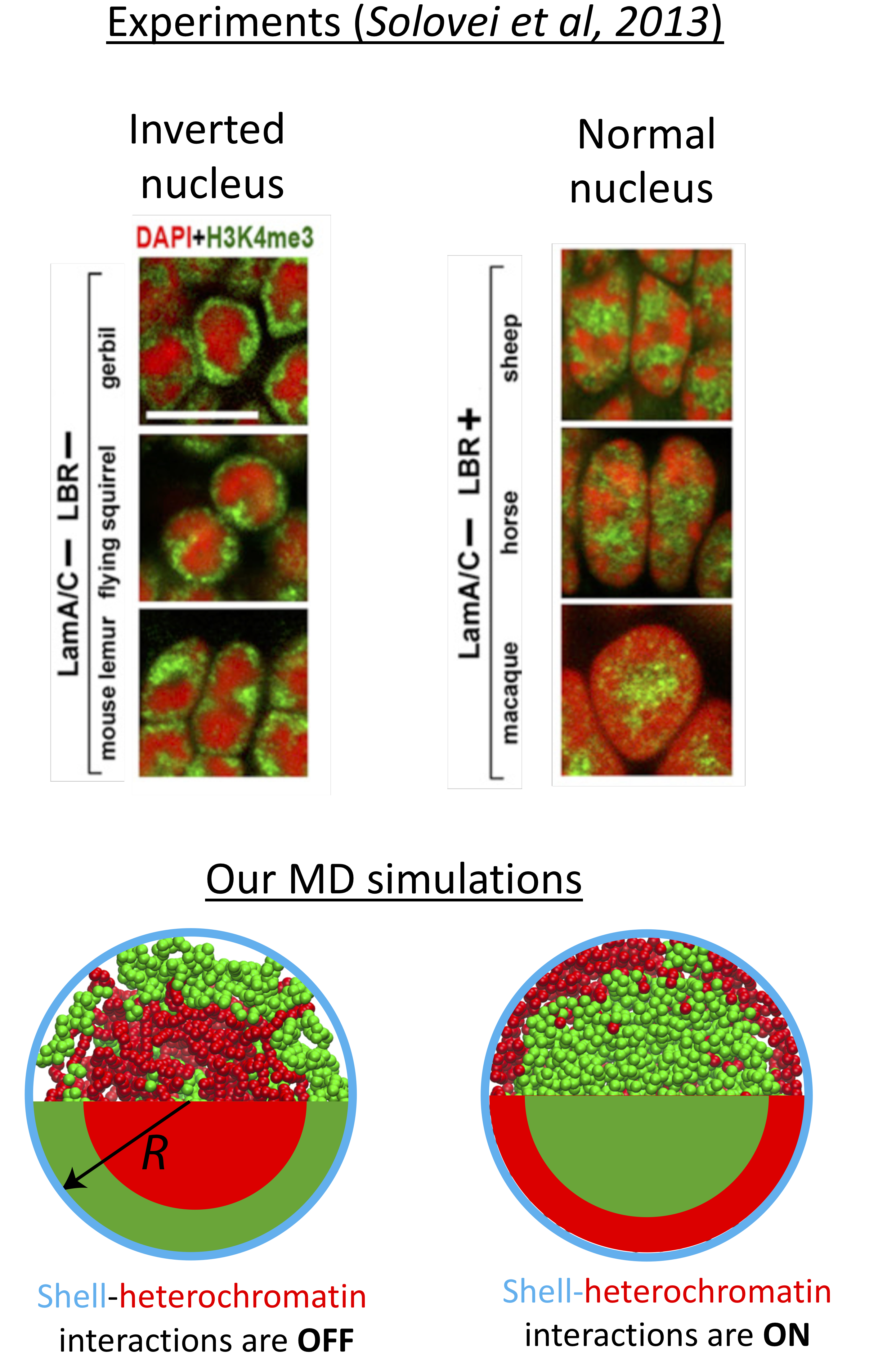
When we look into the organization of genome in the confinement of the cell nucleus, first thing we notice is that distribution of chromatin (a fibrous structure composed of protein-DNA complexes) is not random. In most healthy cells, while gene-passive heterochromatin (red regions in the images above) is found near the interior surfaces of the nuclear envelope, gene-active euchromatin (green) localizes in the nuclear interior. Yet, deviations from regular chromatin distribution are frequent, for example, upon aging, under malignant diseases, or even naturally in rod cells of nocturnal mammals, for instance, when proteins connecting the chromatin to the nuclear lamine is deficient or absent. In order to understand how polymer properties of the long chromatin fiber influences the genome organization, we study the segregation of heterochromatin in cell nucleus by modeling interphase chromosome as diblock ring copolymers confined in a rigid spherical shell by using extensive Molecular Dynamic simulations. Heterochromatin block of each bead-spring chain mildly differs from that of euchromatin via its bending stiffness. Our simulations indicate that in the absence of attractive interactions between nuclear shell and chromatin, the majority of heterochromatin segregates in the nuclear interior due to depletion of less flexible heterochromatin segments from nuclear periphery. The central coalescence of heterochromatin is in accord with experimentally observed positional inversion of heterochromatin in rod cells and exhibits a maximum when heterochromatin is only a factor of two less flexible than euchromatin. Attractive interactions between heterochromatin segments and the bond-crossing (topoisomerase activity) further increase the heterochromatin inversion. The inversion is also found to be independent of the heterochromatin concentration and chromosome number. The inverted nuclei can be reversed by switching on the attraction interactions between the nuclear shell and heterochromatin or by increasing the bending stiffness of the heterochromain. Overall, our results indicate that any molecular phenomenon that can lead to suppressed bending fluctuations along the chromatin could favor nuclear segregation, thus, can have important consequences for gene regulation. Read our preprint.
- How receptor density and reservoir dimensions effect the diffusion process of ligands initially located at their receptors
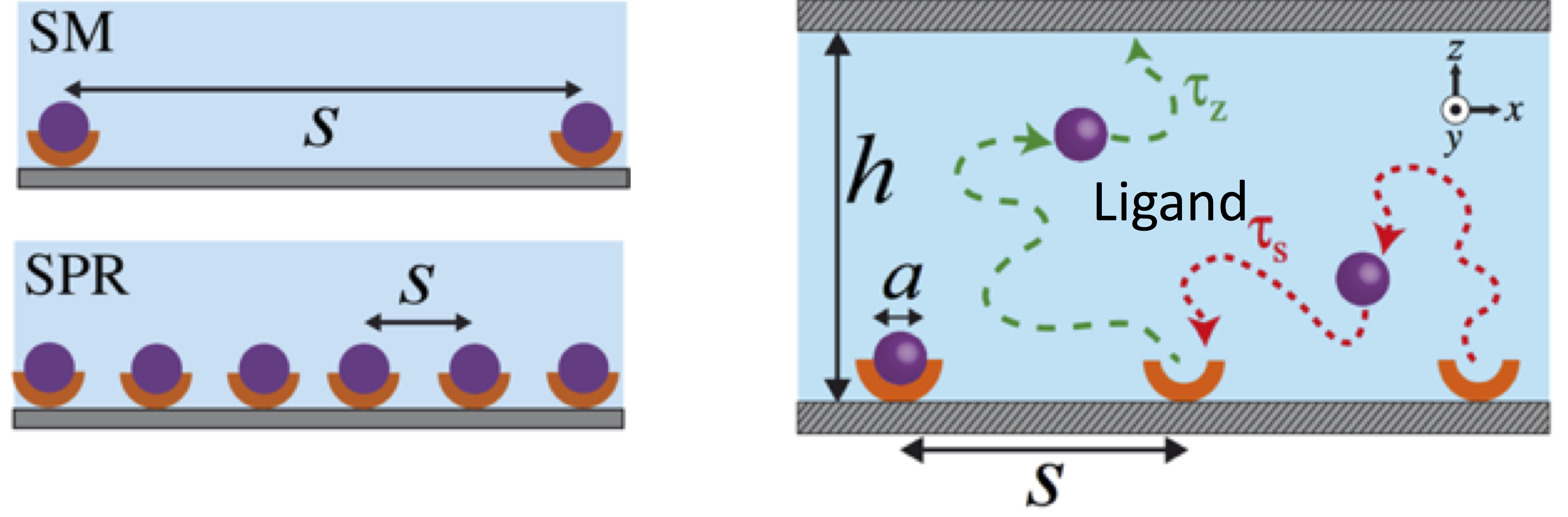
Single-molecule (SM) or SPR (Surface Plasmon Resonance) experiments rely on relaxation of concentration quenches of initially surface-bound molecules into confined reservoirs to determine molecular kinetic rates. Similarly, biological processes such exocytosis, in which small molecules are emitted into the intracellular cleft for cellular communication, can be considered to be a relaxation process of an effective concentration quench. We study a model system closely related to the above cases in which weakly interacting Brownian particles are released from their binding sites into a confined volume by using molecular dynamics simulations and scaling arguments. Our results suggest that the rebinding rate of released particles exhibits various power laws until the confined volume is entirely filled by the particles. Furthermore, the cumulative rebinding rate, which is time integration of the rebinding rate, exhibits a novel plateau behaviour. This plateau is a result of the minimal number of collisions between the binding sites and ligands. Our results can have important consequences for molecular signalling as well as for the interpretation of kinetic measurements of ligand-receptor interactions. Read more on Biophysical Journal.